Image above: Early morning in Loxahatchee Wildlife Refuge.
Employing “-omic” methods to unravel invasion biology
|
Ferns are one of the final frontiers in plant genomics; the first fern genome was published in 2018! We are currently sequencing the genome of the invasive fern Lygodium microphyllum, commonly known as the Old World climbing fern. This invasive species has cost the state of Florida millions of dollars and untold ecological damage to vulnerable ecosystems. We are using a combination of approaches to sequence and assemble the genome of L. microphyllum including short and long-read technologies and chromatin confirmation sequencing. We are working with Fay-Wei Li (Boyce-Thompson Institute/Cornell), Li-Yaung Kuo (National Tsing Hua University), and Trevor Krabbenhoft (University at Buffalo) to sequence this genome. I plan to develop L. microphyllum as a model system for understanding invasive fern biology with the hope of generating resources for applied sciences.
|
Relevant Publications:
- Pelosi, J.A., B.A. Zumwalde, O. Hornych, K. Wheatly, E.H. Kim, and E.B. Sessa. Lygodium japonicum (Lygodiaceae) is represented by a tetraploid cytotype in Florida. The American Fern Journal 113(1): 43-55
Evolution of and variation in life cycle: effects on population and genome evolution
All plants undergo an alternation of generations between gametophyte (haploid) and sporophyte (diploid) life phases, and yet these morphologically and functionally disparate phases share a single genome. Understanding the complexities of the genome and gene expression profiles that facilitate these different phases and how these impact application-based science are some of of my main research aims. I am using several systems to explore these differences, including developing Lygodium microphyllum, Dryopteris spp., and Vittaria appalachiana.
The Appalachian gametophyte, Vittaria appalachiana, is one example of a handful of ferns that have lost the sporophyte portion of their life cycle. We are investigating the genomic consequences of complete asexual reproduction and how extant populations are related. Using a combination of approaches (transcriptomics, population genomics), we aim to unravel some of the mysteries remaining around obligate asexual reproduction in ferns.
The Appalachian gametophyte, Vittaria appalachiana, is one example of a handful of ferns that have lost the sporophyte portion of their life cycle. We are investigating the genomic consequences of complete asexual reproduction and how extant populations are related. Using a combination of approaches (transcriptomics, population genomics), we aim to unravel some of the mysteries remaining around obligate asexual reproduction in ferns.
Relevant Publications:
- Pelosi, J.A., W.B. Barbazuk, and E.B. Sessa. Life without a sporophyte: the origins and genomic consequences of asexual reproduction in a gametophyte-only fern. International Journal of Plant Sciences 184(6): 454-469.
- Pelosi, J.A., E.S. Sorojsrisom, B.A. Zumwalde, and E.B. Sessa. A genome size for the Appalachian gametophyte. The American Fern Journal 113(4): 257-262.
- Aros-Mualin, D., C. Flores-Galván, S. Páez, J.A. Pelosi, E.S. Sorojsrisom, N. Yawn, and J.E. Watkins. In situ observations of gametophytes of six fern species of Costa Rica. The American Fern Journal 113(3): 170-190.
Integrative and Comparative phylogenomics
|
Understanding the relationships among taxa is one of the foundational questions in evolutionary biology. As sequencing costs rapidly decrease, there has been a growing wealth of genomic resources from non-model systems that allows us to address how organisms are related. Using a combination of transcriptomic and target-capture datasets, we have been able to elucidate some of the most difficult to resolve nodes in the backbone of the fern tree of life. Together with Dr. J. Gordon Burleigh at the University of Florida and Dr. Emily Sessa at NYBG, we have begun to unravel some of the processes that shape the evolution of ferns using large genome-wide datasets.
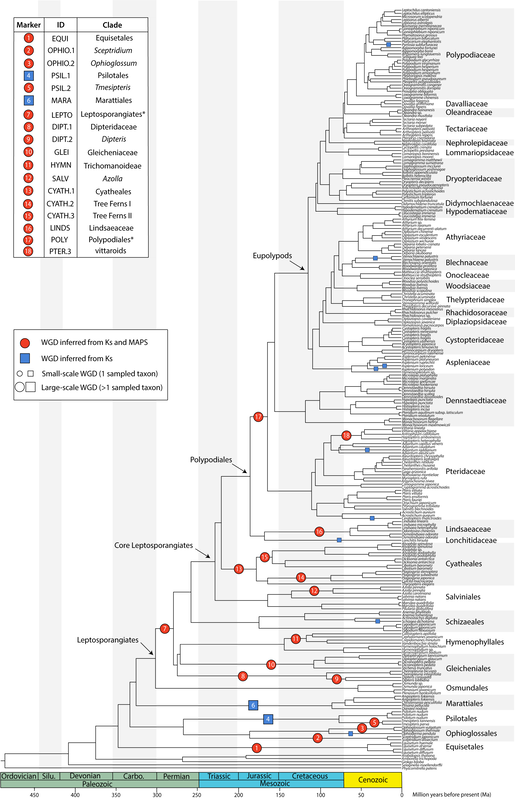
Polyploidy, or whole genome duplication (WGD) is a prominent force in plant evolution, with nearly one-third of speciation events in ferns associated with a change in ploidy. Our recent work has focused on both ancient (paleopolyploidy) and recent/ongoing (neopolyploidy) WGD events and their placement on the phylogeny. We employ bioinformatic and wet-lab approaches to determine ploidy and generate hypotheses of parentage of these putative polyploids. Work has focused on Asplenium, Lygodium, among other ferns. On a larger scale, we are interested in the processes that shape genome evolution following polyploidy and the patterns of gene retention that arise as a consequence. |
Relevant Publications:
- Pelosi, J.A., B.A. Zumwalde, W.L. Testo, E.H. Kim+, J.G. Burleigh, and E.B. Sessa. All tangled up: unraveling phylogenetics and reticulate evolution in the vining ferns, Lygodium (Lygodiaceae, Schizaeales). In revision at the American Journal of Botany.
- Pelosi, J.A., E.S. Sorojsrisom, B.A. Zumwalde, and E.B. Sessa. A genome size for the Appalachian gametophyte. The American Fern Journal 113(4): 257-262.
- Pelosi, J.A., B.A. Zumwalde, O. Hornych, K. Wheatly, E.H. Kim, and E.B. Sessa. 2023. Lygodium japonicum (Lygodiaceae) is represented by a tetraploid cytotype in Florida. The American Fern Journal 113(1):43-55.
- Pelosi, J.A., E.H. Kim, W. B. Barbazuk, and E.B. Sessa. 2022. Phylotranscriptomics illuminates the placement of whole genome duplications and gene retention in fern ferns. Frontiers in Plant Science 13:882441.